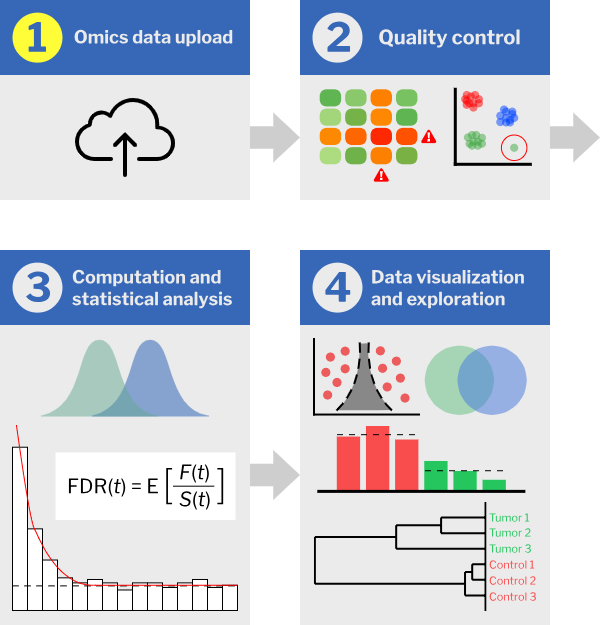
SimpliFi is an online, browser-accessible platform that translates omics data into biological understanding. Users of all skill levels can explore and visualize their data to understand a biological state and form hypotheses. SimpliFi accepts and integrates quantified omics data of all kinds from any method – proteomics, lipidomics, metabolomics, transcriptomics, etc. – and distills data into clean, interactive displays of enriched differentially expressed pathways, GO enrichments, cellular markers, protein-protein interaction networks and molecular-level classifications. UniProt and ChEBI identifiers yield the most utility and all results can be exported with a simple right mouse click.
SimpliFi employs exclusively non-parametric statistics and resampling approaches, modeling biological variability from biological replicates and accounting for observed differences as a function of biological variation, number of samples and observations, and measurement error. This approach correctly addresses non-Gaussian biological distributions and the intensity dependence of omics measurements: 1 vs. 2 counts is not the same as 1000 vs. 2000 counts. SimpliFi uses bootstrapping and resampling approaches to always calculate and display confidence intervals for p-values and fold changes. Missing values are allowed and accounted for. SimpliFi also offers standard statistical tests (t-tests, Wilcoxon, resampling) and obligates batch control and outlier analysis (Pearson's correlations, PCA, hierarchical clustering) prior to enabling further data visualization, exploration or analysis. Users are displayed warnings if batch, or run order effects or outliers are detected.
SimpliFi runs on GPUs to facilitate analysis of even large datasets.